Current projects
Recent genomic sequencing data underline the host obligate nature of the fungus Pneumocystis and provide information about possible requirements for cell culture, the success of which has so far been limited. The use of respiratory tissue from the appropriate host is likely to be decisive. This study will answer the following questions: Does Pneumocystis f. sp. suis (P. suis) show a strict host specificity and a strong nutritional dependency on the porcine host tissue? Is the fungus responsible for only minor tissue lesions?
In preliminary trials, we have attempted various approaches to cultivate pig respiratory tissue. Tracheal explants and monolayer pig lung/airway cells were cultivated, and the latter were transferred to an air/liquid interphase culture. In our study, these systems will be inoculated with P. suis. In case of failure, a cell-free medium cultivation system will be established in parallel, which will enable us to work on further project parts. For inoculation, P. suis organisms will be separated from either bronchial lavage fluid or lungs of immunosuppressed pigs, concentrated and quantified by flow cytometry using commercially available antibodies. In addition, monoclonal anti-P. suis antibodies will be generated by hybridoma technology. P. suis stages and infection-derived tissue lesions will be imaged by transmission electron microscopy. Finally, gene expression of P. suis will be studied by mean of RNA-Seq.
P. suis is one of the less described Pneumocystis species. Cell culture has had only limited success and every contribution to this topic is essential for the entire Pneumocystis community. First steps towards the generation of monoclonal anti-P. suis antibodies by hybridoma technology and their application in cell sorting will be taken to create the basis for future projects. Imaging will contribute to a better understanding of P. suis pathogenesis. Gene expression analysis will unlock basic biological functions and will contribute to understanding the complexity of the fungus-host-interactions.
Involved scientists:
Priv. Doz. Dr. Christiane Weissenbacher-Lang
Dr. Barbara Blasi
Mag. Nicolle Gobbo Oliveira Erünlü
Univ. Prof. Dr. Herbert Weissenböck
Kooperationspartner:innen:
Prof. Dr. Ingrid Walter, Arbeitsgruppe für Histologie, Institut für Morphologie, Veterinärmedizinische Universität Wien
Mag. Natascha Leitner, Arbeitsgruppe für Histologie, Institut für Morphologie, Veterinärmedizinische Universität Wien
Kerstin Mair PhD, Institut für Immunologie, Veterinärmedizinische Universität Wien
Prof. Dr Andrea Ladinig, Universitätsklinik für Schweine, Veterinärmedizinische Universität Wien
Mag. Christian Knecht, Universitätsklinik für Schweine, Veterinärmedizinische Universität Wien
Dr. Joseph A. Kovacs, Critical Care Medicine Department, NIH Clinical Center, Bethesda, Maryland, USA
Dr. Liang Ma, Critical Care Medicine Department, NIH Clinical Center, Bethesda, Maryland, USA
Dr. Ousmane H. Cissé, Critical Care Medicine Department, NIH Clinical Center, Bethesda, Maryland, USA
Lisa R. Bishop, Critical Care Medicine Department, NIH Clinical Center, Bethesda, Maryland, USA
Dr Shelly J. Curran, Critical Care Medicine Department, NIH Clinical Center, Bethesda, Maryland, USA
Project period: 03/2023 – 02/2027
Funding:
FWF – Austrian Science Fund
Using animal models (pig and rat) the effects of mild hypothermia on neuropathological changes after cardiac arrest and reperfusion are investigated. The optimal time point and the best method for induction of hypothermia should be determined.
Involved scientists:
Dr. Sandra Högler
Univ.-Prof. Dr. Peter Schmidt, Dipl.ECVP
Cooperation partners:
Dr. Sandra Högler, Unit of Laboratory Animal Pathology, vetmeduni Vienna
Prof. Sterz, Dr. Wolfgang Weihs, Department für Notfallmedizin, Med.Uni.Vienna
Prof. Sipos, Klinik für Schweine, Vetmeduni Vienna
Catharina Duvigneau, Institut für Biochemie, Vetmeduni Vienna
Funding:
FWF - Austrian Science Fund
Hochschuljubiläumsstiftung der Stadt Wien
AKH
Dead animals submitted for necropsy are frequently the initial event for recognizing and follow-up investigations of emerging diseases (examples: Usutu virus, West Nile virus or Rustrela virus infections).
In some cases the recognition of a novel condition will initiate surveillance programs which provide data on incidence, epidemiology and general relevance of such diseases.
Involved scientists:
Univ.-Prof. Dr. Herbert Weissenböck, Dipl.ECPHM
Priv.-Doz. Dr. Christiane Weissenbacher-Lang
Cooperation partners:
Virology, University of Veterinary Medicine
Universitätsklinik für Schweine
AGES
Friedrich-Loeffler-Institut, Greifswald - Insel Riems
Klinische und vergleichende Neuropathologie, Institut für Tierpathologie, LMU München
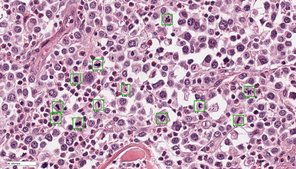
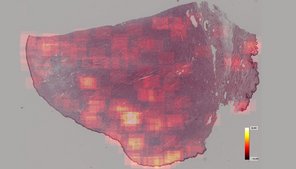
Tissue specimens have been traditionally examined by pathologists using conventional light microscopy. Digital pathology is an emerging field of pathology that uses digital microscopic images and is increasingly replacing traditional light microscopy. Whole slide image (WSI) scanners enable digitization of entire microscopic specimens, which can subsequently be examined by observers (digital microscopy) or be analyzed by computer software (automated image analysis). With this new technology, there are numerous innovative applications for histopathological diagnostic, research and educational purposes. Regarding diagnostics of surgical tissue biopsies, general advantages of digital pathology include improved work flexibility (such as home office), optimized laboratory workflow and increasing accuracy and reproducibility in quantifying histopathological parameters. The latter can be archived by automated image analysis, especially if artificial intelligence is used. Although many diagnostic laboratories are already using digital microscopy, automated image analysis is not routinely applied due to limited availability of software solutions and insufficient knowledge of performance and reliability.
The goals of this research project are to develop innovative software solutions using artificial intelligence for tumor histopathology and validate its diagnostic applicability. This project is conducted in cooperation with research partners from the Technische Hochschule Ingolstadt (workgroup of Prof. Dr. Marc Aubreville), Friedrich-Alexander Universität Erlangen-Nürnberg (workgroup of Prof. Dr. Katharina Breininger), Freie Universität Berlin (workgroup of Prof. Dr. Robert Klopfleisch) and Animal Medical Center, New York (Dr. Taryn A. Donovan). The focus of this project is to optimize current algorithmic solutions in order to allow a broad application in different laboratories, validate different approaches for application of algorithms in a diagnostic setting, evaluate the influence of algorithms on quality and efficiency of diagnosis and prognosis as well as scrutinize advantages and disadvantages / limitations of automated image analysis.